Novel methods capable of quickly identifying single cancer cells while keeping them alive for downstream analyses would enable improved diagnostics and allow clinicians to culture them for personalised drug tests. This article highlights how combining AI and optical microscopy led to an inexpensive and accurate way to classify cancer cells based on their intracellular acidity.

An interesting approach was introduced in 2017 by Hou and co-workers which described a novel simple and inexpensive single-cell pH-based imaging method that allowed for the identification of cancer cells via common pH indicators and UV-vis micro-spectroscopy.
Combining pH-based colorimetric imaging and AI for single-cell classification
Recent advancements in Artificial Intelligence (AI) have enabled novel approaches aimed at extracting information from biomedical images to identify specific phenotypes and aid disease diagnosis. We recently published a work that describes a novel approach where quantitative pH-based colorimetric imaging is combined with AI-based single cell segmentation and classification. This methodology focuses on the use of a pH-sensitive dye, Bromothymol Blue (BTB), as shown in Fig. 1. First, the cell membrane is permeabilised by incubating the cells for 15 minutes with ethanol at 5 per cent concentration. When imaged using a standard optical microscope, upon internalisation of 0.5 mg/mL of BTB, the intracellular environment acquires a characteristic colour based on the unique physiological condition of each cell. Specifically, each type of cell was shown to exhibit a specific colour ‘fingerprint’ consisting of a unique combination of Red, Green, and Blue (RGB) components.
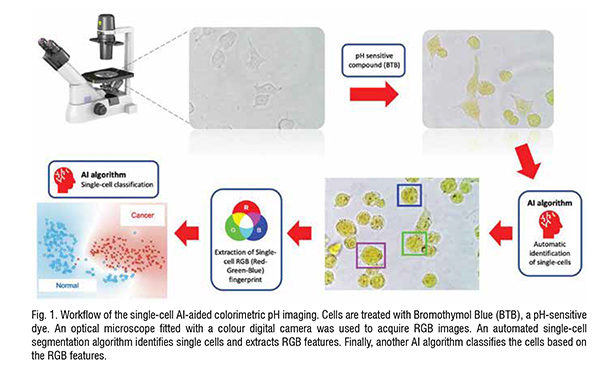
Normal cells (HUVECs) were found to have the lowest levels of pH versus all the cancer cell lines. For example, higher pH was found in the metastatic breast cancer cell line, MDA-MB-231, as compared with the non-cancerous cell line, MCF-10A. These findings are in accordance with past studies.
Moreover, each cell type exhibits a characteristic spatial distribution of the RGB values. This information was also taken into account and used as a feature to automatically identify the different cell types using AI algorithms. The computational classification was performed by comparing an increasing number of different cell types. Singlecell classification accuracy of ~97 per cent was achieved when comparing MCF-10A with MDA-MB-231.
The classification accuracy reached~92 per cent when performing the analysis on three cell lines, including the human pancreatic cancer cell line Mia-aCa-2 to the breast cell lines previously analysed. Finally, adding a fourth cell line to the analysis – the umbilical vein endothelial cells (HUVECs) - a classification accuracy of 92.7 per cent was achieved. The demarcation between the different groups is observed in the t-SNE plot (Fig. 2).
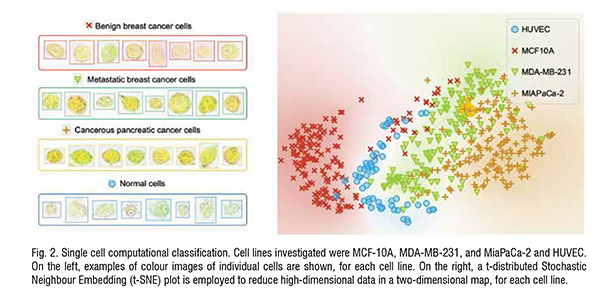
The computational classification was validated using 10-fold crossvalidation for model testing. Further validation was done using both ‘in silico’ co-cultures (where datasets are generated by randomly combining values of the colorimetric features obtained for each cell line) and actual co-cultures.
After going through the analysis pipeline, cells remain viable and can be cultured for several days. Although the use of BTB led to better computational classification accuracy, this method was also successfully proven without the use of BTB, by simply capitalising on the inherent colorimetric differences between cell lines. This is promising and will enable further developments towards a label-free version of this approach. The presented technique could also be used for sub-cellular analyses. In fact, various cellular components have been characterised by different pH levels as reported previously.
Concluding remarks and future perspectives
This method made it possible to discriminate among normal, non-tumorigenic and cancerous cells purely by focusing on their intracellular acidity. The non-invasive nature of this single cell classification methodology makes it extremely easy and useful for identifying various cell types and can be integrated with current screening and diagnostic methodologies. The protocol takes ~35 minutes, including BTB internalisation, image acquisition, image segmentation and classification. Moving forward, a real-time version where single cells in either adherent or suspended state can be quickly detected and individually manipulated. This will offer a less expensive alternative to the current standard flow cytometry-based cell sorting, while maintaining cell viability, which can then be integrated with downstream analysis. To sum up, our method can be used as a fast, inexpensive and accurate tool to diagnose cancer, monitor cancer progression and/or effectiveness of a treatment, and even alerting the risk of a relapse.