Translational Medicine, which aims to improve communication between the basic and clinical sciences, coupled with informatics and semantic technologies will help in creating the next generation healthcare enterprise.
The Web has revolutionised the way people look for information and corporations do business. The Semantic Web, being proposed as the next generation web, builds on the current infrastructure and attempts to give information on the web a well defined meaning. Simultaneously, the life sciences sector is playing host to a battery of innovations triggered by the sequencing of the Human Genome coupled with a more proactive approach to medicine. There is an increased emphasis on disease prevention and wellness of the individual as opposed to disease treatment and management; and significant activity has focused on Translational Research, which seeks to accelerate 'translation' of research insights from biomedical research into clinical practice and vice versa.
The next generation healthcare enterprise - A vision statement
There is a great need to get away from the short-term goals of treating current diseases and conditions, and focus on long-term strategies of enhancing the well-being and quality of life of an individual. In fact, it is a well known fact that adopting the approach of disease prevention will result in reducing the load on current healthcare infrastructure. From this perspective, the vision of the next generation healthcare enterprise may be articulated as follows:
The Next Generation Healthcare Enterprise provides services across the Healthcare and Life Sciences (HCLS) spectrum targeted at delivering optimum wellness, therapy and care. These holistic services cut across biomedical research, clinical research and practice and create a need for the accelerated adoption of genomic and clinical research into clinical practice.
A key consequence of this vision is that not only should the healthcare enterprise meet the current needs of a patient, but also anticipate future needs and implement interventions that can potentially prevent diseases and other adverse clinical events. This could be done by sequencing the genome of the patient and assessing the disposition of a patient towards diseases and adverse clinical events. This creates a need for knowledge sharing, communication and collaboration across the HCLS, which is indeed the underlying goal of Translational Medicine.
Translational Medicine
Translational Medicine aims to improve the communication between basic and clinical science so that more therapeutic insights may be derived from new scientific ideas and vice versa. Translation research goes from bench to bedside where theories emerging from preclinical experimentation are tested on disease-affected human subjects, and from bedside to bench, where information obtained from preliminary human experimentation can be used to refine understanding of the biological principles underpinning the heterogeneity of human disease and polymorphism(s). The products of translational research, such as molecular diagnostic tests are likely to be the first enablers of personalised medicine.
Translation of genomic research into clinical practice
One of the earliest manifestations of translational research will be the adoption of therapies and tests created from genomics research into clinical practice. Consider a patient who suffers a shortness of breath and fatigue in a doctor's clinic. Subsequent examination of the patient reveals the following information:
Information needs and requirements
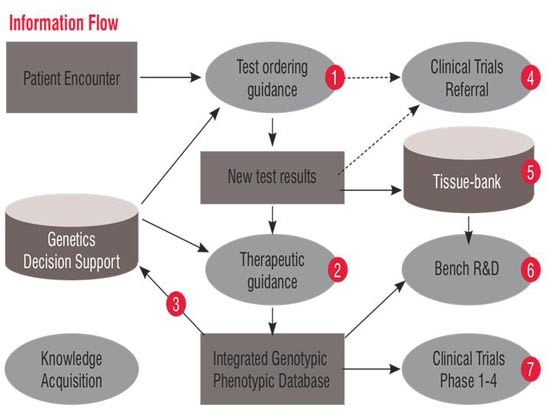
See Figure 1 for information flow to identify various stakeholders and their information needs.
The aggregation of data for identifying patients for clinical trials and tissue banks leads to knowledge acquisition especially to create knowledge bases for decision support. This also helps to map genotypic and phenotypic traits. An enumeration of the information requirements is presented in Table 1.
Service-oriented architectures for translational medicine
Each requirement identified in terms of information items has multiple stakeholders, and is associated with different contexts, such as: (a) domains such as genomics, proteomics or clinical information; (b) activities, such as biomedical research or clinical practice; (c) applications such as the EMR and LIMS; (d) services such as decision support, data integration and knowledge-, provenance-related services.
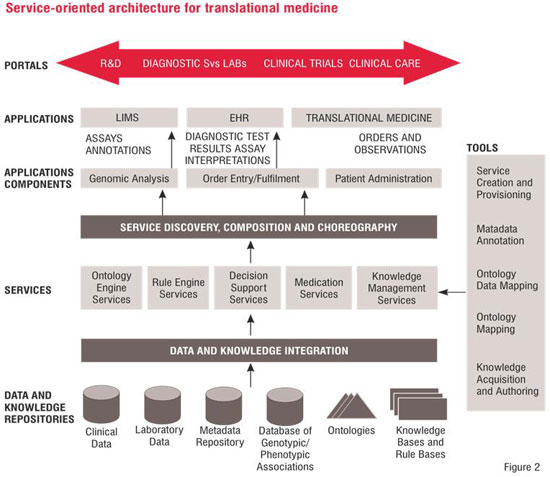
The components of the conceptual architecture, as illustrated in Figure 2, are as follows:
Portals: This is the user interface layer and exposes various personalised portal views for various stakeholders such clinical researchers, lab personnel, clinical trials designers, clinical care providers, hospital administrators and knowledge engineers.
Applications: The two main applications, viz. the EMR system and LIMS are illustrated in the architecture.
Service Discovery, Composition and Choreography: Newly emerging applications are likely to be created via composition of pre-existing services and applications. This component of the architecture is responsible for managing service composition and choreography aspects.
Services: The services that need to be implemented for enabling Translational Medicine applications can be characterised as (a) business or clinical services, e.g. medication and clinical decision support services; and (b) infrastructural or technological services, e.g. ontology and rule engine services.
Data and Knowledge Integration: This enables integration of genotypic and phenotypic patient data and reference information data, which could enable clinical care transactions and discovery of promising drug targets. Examples of knowledge integration would be merging of ontologies and knowledge bases to be used for clinical decision support.
Data and Knowledge Repositories: These refer to the various data, metadata and knowledge repositories that exist in healthcare and life sciences organisations. Some examples are databases containing clinical information and results of laboratory tests for patients. Metadata related to various knowledge objects (e.g. creation data, author, category of knowledge) are stored in a metadata repository.
Data and information integration
We will describe this with the help of an example, an approach for data integration based on semantic web specifications such as the Resource Description Framework (RDF) and the Web Ontology Language (OWL), to bridge clinical data obtained from an EMR and genomic data obtained from a Laboratory Information Management System (LIMS). The first key step in semantic data integration is the definition of a domain ontology spanning across multiple domains; or creation of inter-ontology mappings across multiple ontologies that reflect different perspectives e.g. research and practice a given (clinical) domain.
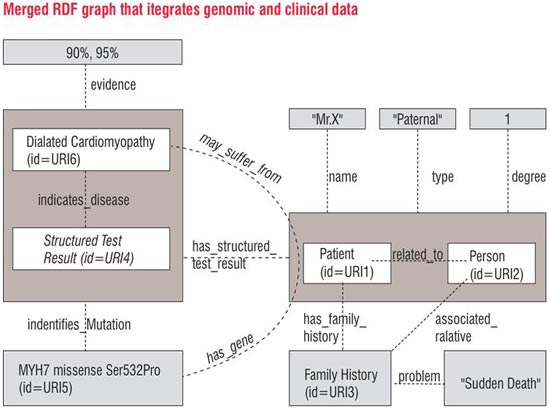
The RDF graphs illustrated in Figure 3 represent clinical data related to a patient with family history of Sudden Death. Nodes (boxes) corresponding Patient ID and Person ID are connected by an edge labelled related_to modelling the relationship between a patient and his father. The name of the patient ('Mr. X') is modelled as another node, and is linked to the patient node via an edge labelled name. Properties of the relationship between the patient ID and person ID nodes are represented by reification (represented as a big box) of the edge labelled related_to and attaching labelled edges for properties such as the type of relationship (paternal) and the degree of the relationship.
Genomic data related to a patient evaluated for a given mutation (MYH7 missense Ser532Pro) is illustrated. Nodes (boxes) corresponding to Patient ID and Molecular Diagnostic Test Result ID are connected by an edge labelled has_structured_test_result modelling the relationship between a patient and his molecular diagnostic test result. Nodes are created for the genetic mutation MYH7 missense Ser532Pro and the disease Dialated Cardiomyopathy. The relationship of the test result to the genetic mutation and disease is modelled using the labelled edges identifies_mutation and indicates_disease respectively. The degree of evidence for the dialated cardiomyopathy is represented by reification (represented as boxes and ovals) of the indicates_disease relationship and attaching labelled edges evidence1 and evidence2 to reified edge. Multiple confidence values expressed by different experts can be represented by reifying the edge multiple times.
The end user previews them and specifies a set of rules for linking nodes across different RDF models. These simple rules may include: merging of nodes that have same IDs or URIs, introduction of new edges based on pre-specified declarative rules specified by subject matter experts and informaticians. New edges that are inferred (e.g. suffers_from) may be added back to the system based on the results of the integration. Sophisticated data mining that determines the confidence and support for new relationships might be invoked. This integration process results in generation of merged RDF graphs as shown in Figure 4.
Conclusion
There is a growing realisation that Healthcare and Life Sciences is a knowledge-intensive field and the ability to capture and leverage semantics via inference or query processing is crucial for enabling translational medicine. Given the wide canvas and the relatively frequent knowledge changes that occur in this area, we need to support incremental and cost-effective approaches to support 'as needed' data integration. Personalised / Translational Medicine needs Semantic Web technologies to be implemented in a scalable, efficient and extensible manner.
AUTHOR BIO
Vipul Kashyap is a Senior Medical Informatician in the Clinical Informatics Research & Development group at Partners HealthCare System and is currently the Chief Architect of a Knowledge Management Platform that enables browsing, retrieval, aggregation, analysis and management of clinical knowledge across the Partners Healthcare System. Vipul has worked on semantics and knowledge-based approaches for information and knowledge management.